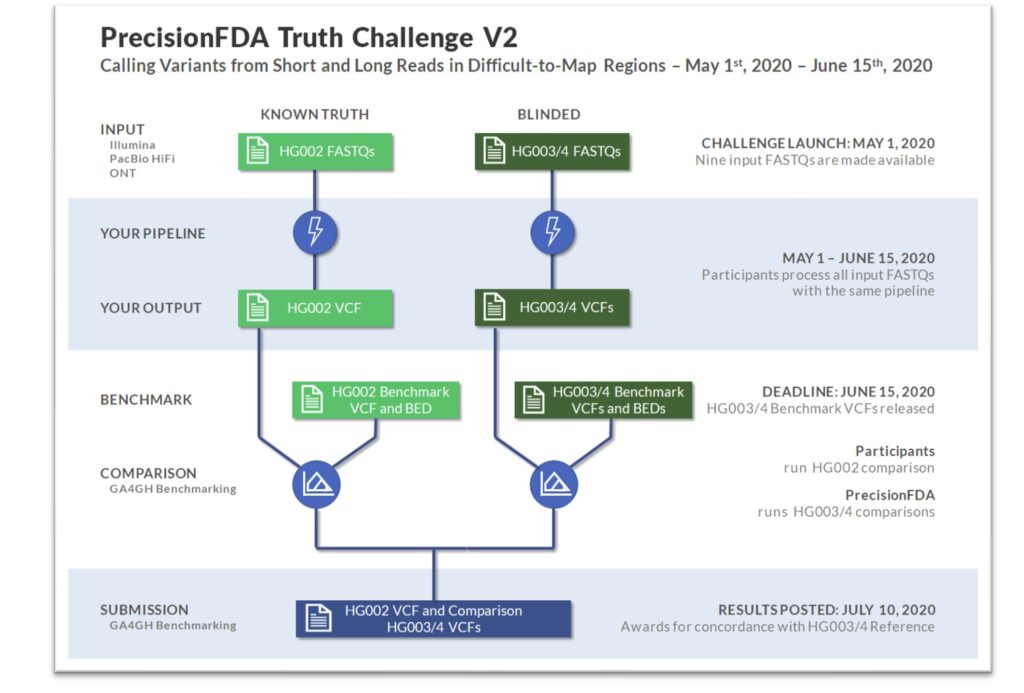
Sentieon wins the precisionFDA Truth Challenge V2 and launches…
Background
In 2016, PrecisionFDA hosted the first Truth Challenge for variant detection. Sentieon, a startup company, participated for the first time and won four out of six categories, including the most prestigious “Best Overall Performance” and “Best Reproducibility,” earning industry recognition.

Four years later, both genome sequencing platforms and analysis software have made significant progress, and the HG00x series of standard samples have undergone iterative updates, covering more and more genome regions. Leveraging the opportunity of Genome in a Bottle releasing the new version 4.1 standard samples, PrecisionFDA hosted a new Truth Challenge V2, encouraging participating teams to challenge the “difficult regions” in the genome using newly developed analysis software combined with long-read sequencing.
Truth Challenge V2
All participating teams in this challenge will use the publicly available HG002 V4.1 standard sample to debug and train their analysis models, and then calculate and compare the accuracy on the HG003 and HG004 standard samples. For each sample, the platform provides Illumina data with ~35x depth, PacBio HiFi data with ~35x depth, and ONT data with ~50-80x depth. Participating teams can choose to analyze only one type of sequencing data or to comprehensively analyze multiple types of data. The final evaluation is divided into three categories, including all genome regions and two additional categories, “difficult-to-map” and “MHC,” which are particularly challenging regions.
After six weeks of workflow development and debugging, a total of 20 teams submitted 64 results, including 24 Illumina data results, 17 PacBio data results, and 20 results from multi-platform integrated analysis. As shown in the figure below, the best performance was achieved by the multi-platform integrated analysis results, followed by PacBio data, with Illumina data in the middle and ONT data analysis results in last place.
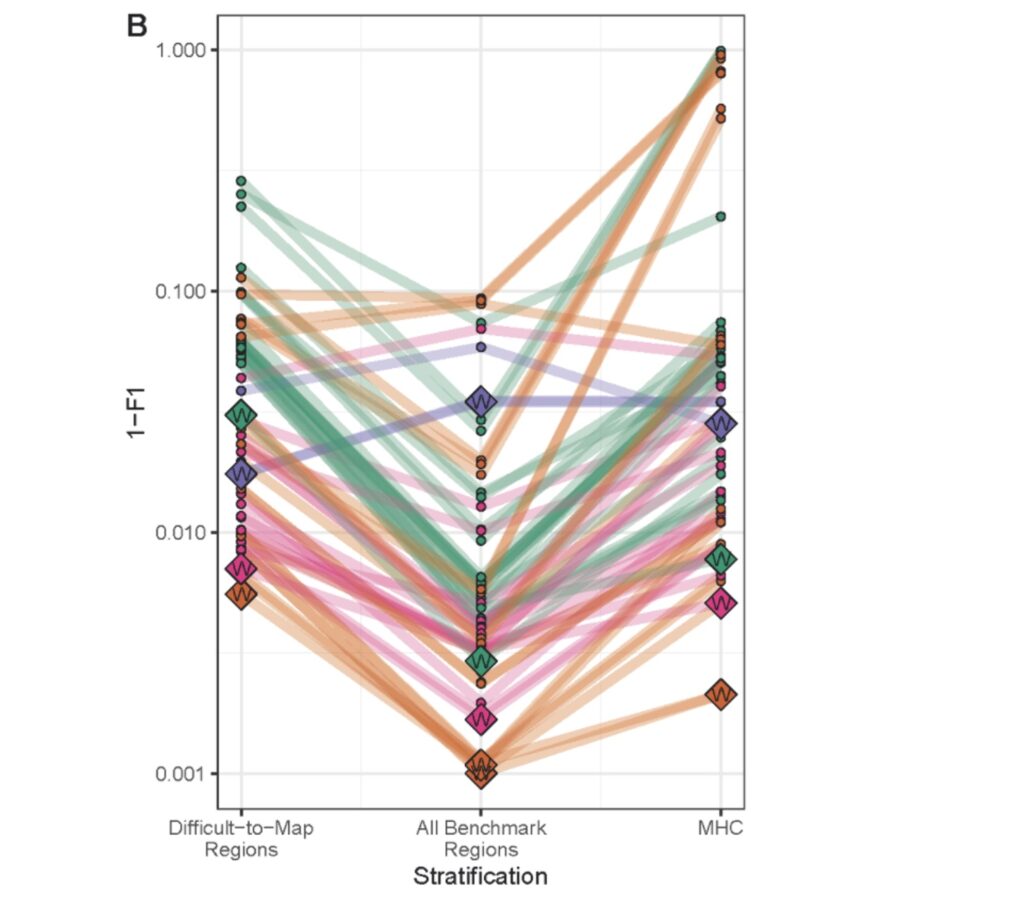
We are pleased to announce that Sentieon has won four championship titles in this challenge, ranking first among all participating teams, including the accuracy championship in the most complex data type category, “All Benchmark Regions – MULTI,” which includes all genome regions from multiple sequencing platforms.
As a team dedicated to bioinformatics algorithm development, this is the sixth time Sentieon has won a PrecisionFDA challenge, maintaining our undefeated record in this series of challenges.

The development and maintenance of a genome sequencing reference standard is crucial for the development of the entire genetic disease research and clinical practice technology process. The upstream sample library preparation, midstream sequencing, downstream secondary analysis, and variant annotation correction and optimization all rely on the accuracy information provided by the reference standard alignment. However, for many years, the available reference standards only covered a limited number of “high-confidence” regions in the entire genome. The remaining regions were excluded from the high-quality reference standard regions due to extreme GC content, simple base repeats, and other reasons that make it difficult to obtain high-quality sequencing sequences. This limitation has caused trouble for many scientific or clinical projects, especially for more advanced research on functional areas outside of the gene exome, such as MHC research.
Compared with GATK, Sentieon’s DNAscope software has more excellent underlying algorithms and a more powerful local reassembly module, which is particularly suitable for processing more complex genomic regions. In the MHC sub-challenge of this challenge, Sentieon won the PacBio and Multi championships, fully demonstrating this point.
The rapid popularization of third-generation sequencing machines PacBio and ONT, especially the production of PacBio Hifi high-accuracy data, has provided powerful tools for cracking these “low-confidence” regions. Long (~15Kb) reads can easily span simple repeat regions and, combined with the accuracy of comparable second-generation sequencing data, can detect multiple types of mutations including SNPs, Indels, and SVs.
Sentieon’s machine learning variant detection software DNAscope was initially developed for Illumina sequencing data types, but the machine learning framework gives the software the potential to adapt to new sequencing platforms. Prior to this challenge, DNAscope had released models adapted to Huada’s sequencing machines, and now it has added a new PacBio-adapted model, with additional models for other sequencing platforms to be released in the future.