Sentieon empowers Oracle Cloud to open the era of…
Recently, Oracle Cloud Infrastructure (OCI) announced the latest achievement in genome applications on its cloud computing platform through a blog post. By using the Sentieon analysis software on OCI, the computing cost for processing genetic sequencing data for users has been significantly reduced, with the on-demand computing cost for whole genome analysis reduced to below $1.

According to the author David Chen (currently serving as OCI Master Principal Cloud Architect), the purpose of this project is to explore how to utilize OCI’s unique hardware resource allocation and optimization mechanisms on a cloud computing platform, coupled with the latest ARM and x86 processors, and the efficient computing efficiency of Sentieon software, to help end users achieve the best balance between efficiency and economy.
With the rapid development of genome sequencing applications, deploying the analysis process on the cloud has become a popular solution to address the pain points of speed and cost brought by massive data. However, in practice, users often directly use unoptimized software and hardware configurations, frequently needing to pay over $10 for each genome analysis. Therefore, choosing suitable hardware instances, setting matching memory, storage, and input/output to achieve cost and speed optimization are worth exploring. For example, are the latest ARM processors more cost-effective than x86 in terms of computation costs? What is the advantage of ARM compared to x86? How does the efficiency and cost of genome analysis process on ARM processors? The purpose of this evaluation project is to provide the best practice for these questions. It is worth noting that the author emphasizes that all costs are calculated based on on-demand pricing, not spot instance pricing. Although on-demand instances are expensive, they provide a guarantee for long-term use and can provide higher practical reference value.
In terms of the analysis process, OCI chose Sentieon DNAseq (v202112.01) as the software solution. As the commercial accelerated process for the current gold standard BWA-GATK best practice, DNAseq can speed up 5-20 times while providing matching results, and provides ARM and x86 compatible instruction set versions.
OCI Instance Benchmark Environment
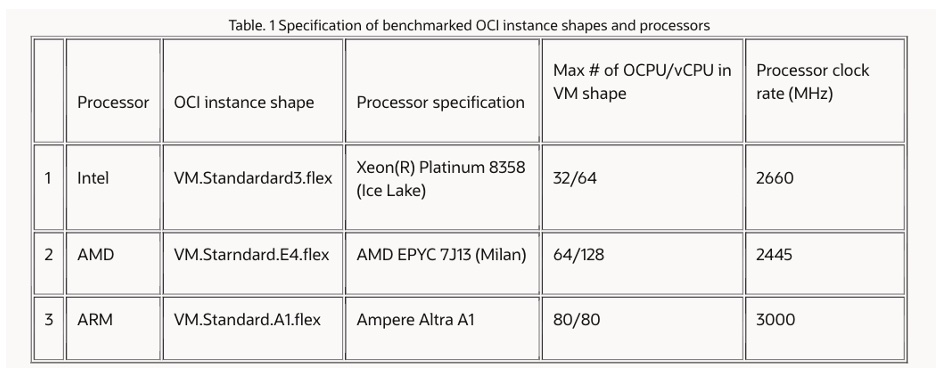
OCI provides different instances based on Intel, AMD, and ARM processors. The author chose three of them (Table 1). Compared to other cloud computing service platforms, one of the advantages of OCI is that it allows users to flexibly specify the required CPU thread number, memory number, storage, and input/output to achieve the best adaptation and optimal usage cost with the application process.
Benchmark Setup
The author focused on the best practice workflow for whole genome embryo analysis, using the Sentieon DNAseq workflow to provide matching results consistent with the best process. The author downloaded and analyzed a total of 7 whole genome data sets, and because the analysis time and cost of all data sets were similar, the author selected only two data set results for display. These two data sets are 30x HG002 standard samples, PCR-free libraries, sequenced by HiSeqX and NovaSeq platforms, respectively.
The GRCh38 reference genome was selected for this analysis process, which includes primary contigs and additional decoy contigs, but not ALT contigs or HLA genes. Compared to the previous GRCh37 version, the current GRCh38 reference genome significantly improves the integrity and accuracy of the sequence and is widely adopted as an industry standard. The analysis process takes FASTQ files as input and VCF files as output, including multiple steps such as alignment, sorting, deduplication, BQSR, and variant detection.
Benchmark Results
Based on the three physical instances, the author created a total of 11 test instances according to different thread numbers, memory, and storage resource requests, including three based on Intel processors, four based on AMD processors, and four based on ARM processors. Specific configurations and computing cost details are shown in the table below.
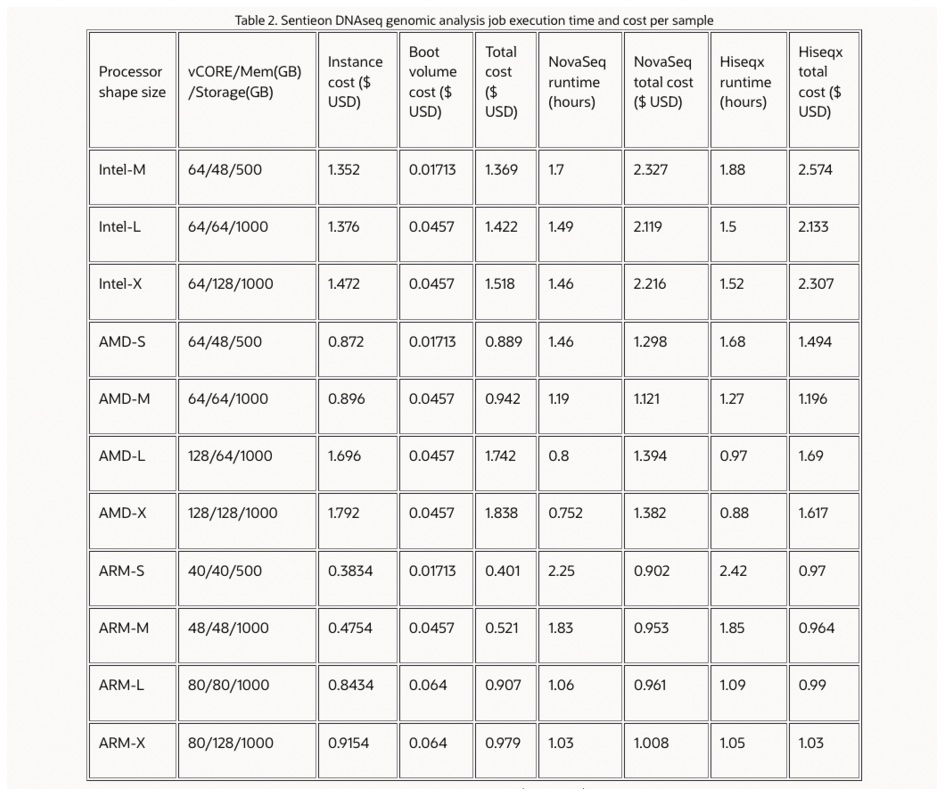
Note 1: The hourly cost per instance comes from the OCI product cost estimator. In addition, the corresponding storage cost of 500G ($0.0171/hour) or 1000G ($0.0457/hour) is added. The input/output selection for all instances is “balanced block volume (VPU:10) with IOPS target of 25,000 and throughput target of 240 or 480 MB/s”.
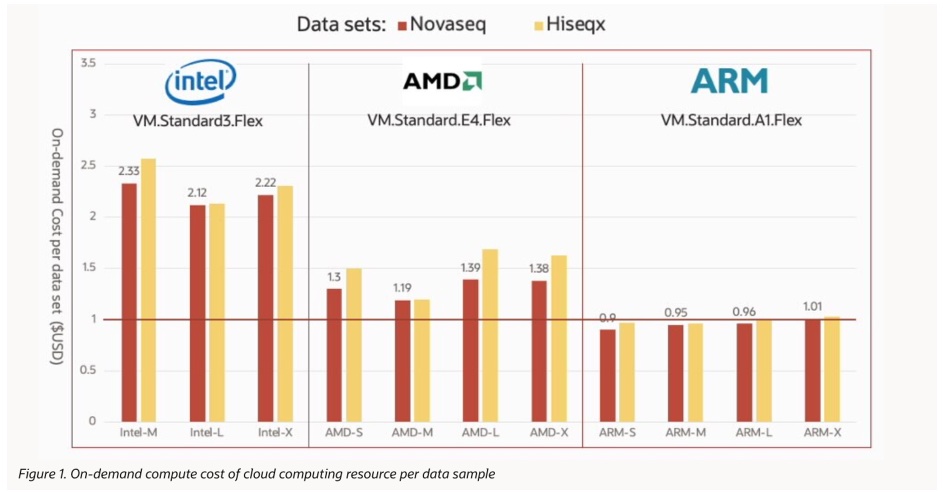
The test results show that, with the full performance optimization of Sentieon on ARM processors, the overall analysis cost of DNAseq for 30x WGS is less than $1 on most ARM instances on OCI, with the lowest cost of $0.9 on the ARM-S instance for processing NovaSeq 30x WGS. In addition, the computational cost of the NovaSeq dataset is slightly lower than that of the HiSeqX dataset.
Analysis of the balance between performance and cost
It is not enough to consider only the economy, users also want to make a comprehensive analysis between speed and economy. From the speed and cost display chart in the figure below, we can see intuitively that small ARM instances have the best economy, while large AMD instances have the best speed. Large ARM instances (80 cores) demonstrate a more balanced economy and speed, and can complete the analysis of the whole genome in 1 hour with a computation cost of 1 dollar.
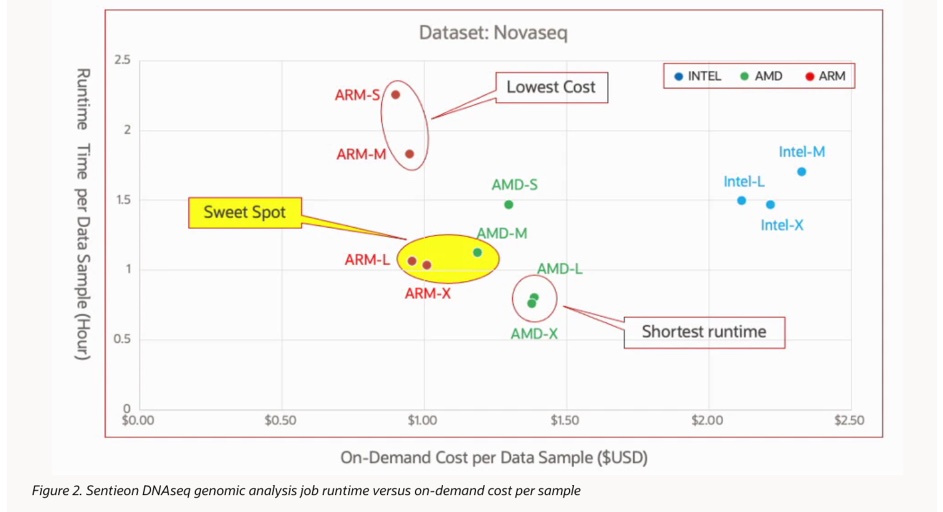
Note that in the same number of cores, the speed of Intel and AMD instances is still relatively similar. In this test, the available CPU cores for the Intel instance were limited, and high-threaded performance with more than 64 threads was not tested like other CPU types. At the same time, on the OCI platform, Intel’s pricing per thread is slightly higher than other CPUs. However, the author announced that the next phase of the evaluation will focus on Sentieon distributed computing, with a particular focus on the performance of Intel instances and their unique RDMA network memory.
Computational Resource Monitoring
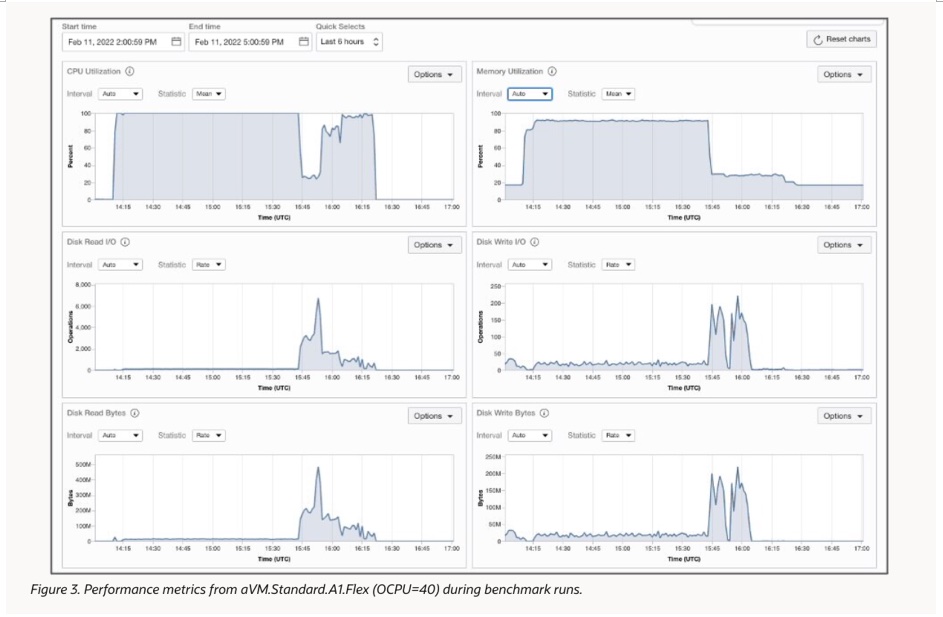
OCI’s excellent scalability ensures that users can find the optimal balance between speed and cost when configuring full genome and other analysis processes. At the same time, OCI’s control panel provides resource monitoring tools to help users confirm whether the computational resources called match the process requirements. The figure below shows the performance of computational resources when running full genome analysis on the VM.Standard.A1.Flex instance.
It should be noted that the speed of Intel and AMD instances under the same number of cores is quite similar. In this test, the Intel instance was limited by the smaller number of available CPU cores and did not test the high-threaded performance of more than 64 threads like other CPU types. At the same time, on the OCI platform, Intel’s pricing per thread is slightly higher compared to other CPUs. However, the author has announced that in the next phase, the evaluation based on Sentieon distributed computing will be carried out, focusing on the performance of Intel instances and its unique RDMA network memory.
Resource monitoring
OCI’s excellent resource scalability ensures that users can find the best speed and cost balance when configuring the entire genome and other analysis processes. At the same time, the OCI platform’s control panel also provides resource monitoring tools to help users confirm whether the computational resources called match the process requirements. As shown in the figure below, the performance of computational resources when running whole-genome analysis on the VM.Standard.A1.Flex instance is displayed.
In the analysis process of the entire genome, the alignment and variation detection steps mainly rely on CPU calculations, while sorting and deduplication are steps with heavier input/output requirements. The resource monitoring results show that Sentieon DNAseq has achieved nearly 100% CPU utilization in most time periods, only slightly lower in sorting and deduplication steps, when the input/output peak also approached the set upper limit of 240MB/s. In addition, the memory utilization rate remained around 90% most of the time. Sentieon DNAseq has efficiently and fully utilized all available hardware resources provided by the OCI platform with deep optimization for different processors, achieving the best economy.
Furthermore, it was found that if the storage limit is increased from 500GB to 1000GB and the input/output limit is increased from 240MB/s to 480MB/s, the steps with larger input/output requirements will be accelerated, and additional 5-8 minutes of acceleration time can be obtained in most instances.
Discussion
CPU
In addition to single-instance computing, Sentieon DNAseq can also parallelly process single samples on multiple instances through distributed computing to obtain further acceleration. The author stated that in the next blog, the performance of DNAseq in distributed computing will be evaluated and summarized. In addition, this evaluation confirmed that the analysis process can use the hyper-threading function of the x86 platform. Although ARM processors do not support hyper-threading, the high economy makes ARM able to provide more physical cores to make up for it.
Memory
Sentieon DNAseq’s memory management is very efficient, and only less than 10GB of memory is required for steps other than alignment. This makes it more convenient for users to analyze larger amounts of sample data because expensive large memory instances are not required. In addition, it was noticed that in the case of sufficient memory, Sentieon’s alignment step can save more intermediate files in memory to obtain additional speed benefits, but this benefit will reach saturation at 128G of memory.
Input/output
To be as close as possible to the actual production environment, the author output typical necessary output files, including BAM, VCF, QC, etc., with an output file size of about 90GB-120GB in this evaluation. All instances in this evaluation chose “single boot volume” as the storage system. In addition, the author confirmed that when the storage and input/output are increased from 500GB (240MB/s) to 1000GB (480MB/s), the input/output speed of the entire process has also been improved to some extent.
Analysis throughput and scalability
The author defines “throughput” as the number of 30x whole genome samples that can be processed per day. The following graph shows the relationship between throughput and the number of threads used for different instances. It can be seen that the throughput in this test ranged from 10 genomes/day (ARM-S) to 32 genomes/day (AMD-X).
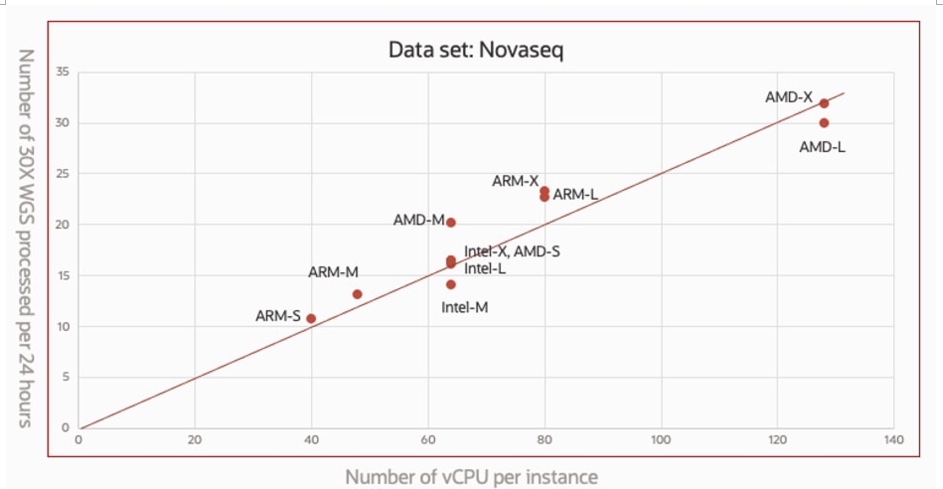
The figure also demonstrates the excellent scalability of Sentieon DNAseq on the OCI platform. As we can see, the overall throughput is approximately linearly related to the number of available threads, proving that the OCI platform can provide maximum computational resources for different CPU architectures. Meanwhile, Sentieon DNAseq’s algorithm optimization and instruction set optimization for different CPU architectures efficiently utilize all available hardware resources.
Conclusion:
- The latest ARM instances provided by the OCI platform are capable of heavy-duty HPC workloads, including genome-wide analysis, which requires high computational performance. As shown in the scalability chart (Fig 4), ARM instances can provide the same computational capacity as x86 instances with the same number of threads, solely through physical cores without hyper-threading. Based on the excellent performance and cost-effectiveness of ARM processors, OCI’s ARM instances can complete FASTQ-VCF processing of 30x whole-genome data within an hour, bringing on-demand computing costs of less than $1 for end-users.
- The OCI platform’s various computing resources can be finely tuned to precisely match the requirements of tasks, reducing resource waste and lowering usage costs. In this evaluation, the authors found the balance between the speed and cost of whole-genome analysis, bringing direct cost reduction to end-users in the life science and health industries. These users typically have to pay more than $10 per genome for the same type of analysis, while the OCI platform can bring enormous value to these professional users.
- The Sentieon DNAseq pipeline matches the industry gold standard BWA-GATK best practices, with a speed increase of 5-20 times. DNAseq is optimized for x86 or ARM processors, which can be directly used on these two platforms, eliminating the additional work of user installation and configuration. The efficient utilization of computing resources by the DNAseq pipeline is an indispensable aid for the OCI platform to achieve one-hour, $1 genome analysis.
“On-demand configuration, pay-as-you-go.” This is the core cost function provided by Oracle Cloud Infrastructure.
This evaluation proves OCI’s vision of providing the lowest computing costs on public cloud platforms. The Sentieon pipeline has been deployed in OCI’s marketplace, providing 30 days of free use and $1,000 in pre-charged amounts for applicants from academia. Welcome to try it out!